INTRODUCTION
MATERIALS AND METHODS
Plant material
Detection of seed lipoxygenases: Colorimetric analysis
Whole-genome re-sequencing
DNA extraction and KASP assays
Statistical analysis
RESULTS
Genetic locations of lipoxygenase genes
Analysis of lipoxygenase in 1,001 soybean accessions
Whole-genome re-sequencing
Lox1 gene mutation
Detection of mutations in lipoxygenase genes using KASP markers
DISCUSSION
INTRODUCTION
Soybean (Glycine max L.) is a globally recognized and important crop that contains oils and proteins (Farhad et al., 2010). Although soybeans are primarily used as animal feed, they are also widely used in food, feed, and industrial applications, such as biodiesel production (Clemente & Cahoon, 2009). However, soybeans contain lipoxygenase, an aromatic nutrient that produces a grassy flavor in food (Gerde & White, 2008). Lipoxygenase catalyzes the oxidation of polyunsaturated fatty acids such as linoleic acid (18:2) and linolenic acid (18:3), resulting in the production of oxidation products, which are medium-chain aldehydes and alcohols associated with the development of undesirable grassy and bitter tastes in soy products (Arai et al., 1970; Sasaki & Chiba, 1983; Wolf, 1975). Various treatments, such as heat, extraction with organic solvents, and decomposition of the beany flavor by aldehyde dehydrogenase, have been studied to reduce these problems during soybean processing (Kitamura, 1984). However, these treatments are expensive and are not entirely satisfactory for food materials because of the poor solubility of soybean proteins caused by heat treatment (Kitamura, 1984). Therefore, an effective solution is to use soybean lines lacking seed lipoxygenase to enhance the suitability of soybeans as food.
Lipoxygenases, a class of enzymes widely distributed in the biological world, are found in plants, fungi, and animals (Brash, 1999). In mature soybean seeds, there are at least three lipoxygenase isoenzymes: Lox1, Lox2, and Lox3 (Axelrod, 1981). Genetic analysis has revealed that the Lox1 and Lox2 loci are closely linked, sharing 81% sequence similarity on chromosome 13 (Siedow, 1991). Moreover, mutant alleles of Lox1 and Lox2 have been found in independent germplasms, indicating repulsion-phase inheritance (Davies & Nielsen, 1986; Hildebrand & Hymowitz, 1982; Kitamura et al., 1985). In contrast, the Lox3 locus on chromosome 15 segregates independently of Lox1 and Lox2, suggesting a distinct mode of inheritance (Davies & Nielsen, 1986; Hajika et al., 1992; Kitamura et al., 1985). Consequently, Lox1 and Lox2 are closely linked and coinherited, whereas Lox3 follows an independent inheritance pattern (Siedow, 1991). Functionally, Lox3 serves as the primary isozyme and demonstrates the highest catalytic proficiency in secondary reactions. It efficiently promotes these reactions regardless of whether they occur under aerobic or anaerobic conditions. Conversely, Lox1 primarily participates in secondary reactions at low oxygen levels or anaerobic environments, whereas Lox2 exhibits minimal secondary product formation under various conditions. This functional characterization underscores the distinct roles of these lipoxygenase isoenzymes (Siedow, 1991).
Early genetic studies demonstrated that the lipoxygenase deficiency is a single Mendelian recessive trait caused by a recessive allele (Hildebrand & Hymowitz, 1982; Kitamura et al., 1983; Kitamura et al., 1985). The presence of the two mutations in Lox1 may be due to the molecular basis of the genotypes identified in previous studies. The Lox1-a allele (PI408251) has a 74-bp deletion at position 2,752 of exon 8, which introduces a stop codon and terminates protein synthesis after 524 amino acids. A nonsense mutation (C2880A) in the Lox1-b allele (PI133226) results in a S568STOP change (Lenis et al., 2010). A missense mutation involving the substitution of T with A in the Lox2 allele has previously been shown to alter one of the conserved histidines ( CAT to CAA) (Wang et al., 1994). PI 205085 and PI 417458 (Lox3-a) exhibited a single guanine deletion within a series of five guanine repeats in the Lox3 allele, starting at position 97 relative to the start codon. This deletion causes a frameshift at position 101, ultimately leading to the premature truncation of the protein at position 42 (Lenis et al., 2010).
Colorimetric assay is the most commonly used method for detecting seed lipoxygenase (Kitamura et al., 1983; Narvel et al., 2000; Suda et al., 1995). This assay takes advantage of the fact that lipoxygenases can be distinguished from each other based on various biochemical properties, such as reaction pH optima, substrate specificity, and protein isoelectric point (Mack et al., 1987). Lox1 activity is optimal at pH levels centered around 9.0, whereas Lox2 activity exhibits a sharp increase around pH 6.5, and Lox3 displays a broad activity range centered around pH 7 (Axelrod, 1981). These assays are valuable tools for rapid screening of seed lipoxygenases in soybean breeding programs focused on selecting varieties with no unpleasant odor or taste.
Colorimetric analysis is subjective, prone to error, and cannot identify heterozygotes. However, the use of molecular markers designed to identify Lox-deficient soybeans enhanced the precision and efficiency of selection. The purpose of this study was to identify soybean accessions in a Korean soybean germplasm collection lacking Lox1, Lox2, and Lox3. Additionally, we aimed to develop a KASP analysis system capable of efficiently identifying soybeans lacking lipoxygenase genes. This approach holds significant promise for simplifying and enhancing breeding efforts aimed at producing soybean varieties with no unpleasant odors or tastes. The use of KASP markers can expedite the selection process and enhance the overall quality of soybean crops.
MATERIALS AND METHODS
Plant material
A total of 1,001 soybean accessions were obtained from the National Agrobiodiversity Center, National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea, and used to analyze the three lipoxygenase isozymes.
In the summer of 2020, Daepung 2 (lipoxygenase wild type) was crossed with IT160160 and IT276392 (null Lox1) at Chonnam National University, Korea. The F1 seeds were advanced to F2 plants in a greenhouse during the winter of 2020–2021, and the F2 seeds were subsequently planted at Chonnam National University in the summer of 2021. In the present study, we generated 169 individual F2:3 populations for Daepung 2 × IT160160 and 147 individual F2:3 populations for Daepung 2 × IT276392.
PI86023 (null Lox2), PI205085 (null Lox3), and PI417458 (null Lox3) were obtained from the USDA Germplasm Resources Information Network (GRIN) (http://www.ars-grin.gov) and used to test the KASP markers. In addition, ‘Jinpumkong 2’ (Kim et al., 1997) was used as a control variety for a triple lipoxygenase mutant.
Detection of seed lipoxygenases: Colorimetric analysis
Lox1, Lox2, and Lox3 were detected using a slightly modified colorimetric assay based on a previously described method (Suda et al., 1995). The lipoxygenase solution was extracted after reacting 50 mg of dried seeds with 300 μL of seed protein extraction buffer and allowing it to incubate for 6 hours. In the case of Lox1, 70 μL of substrate solution (200 mM sodium borate buffer at pH 9.0, 0.1 mM methylene blue, 10 mM sodium linoleate, and distilled water) was mixed with 30 μL of the lipoxygenase solution. For Lox2, 80 μL of substrate solution (200 mM sodium phosphate buffer at pH 6.0, 0.1 mM methylene blue, 10 mM sodium linoleate, 12.5% acetone, and 200 mM DTT) was mixed with 20 μL of the lipoxygenase solution. For Lox3, 80 μL of substrate solution (200 mM sodium phosphate buffer at pH 6.6, 10 mM sodium linoleate, b-carotene [50% saturation], and distilled water) was mixed with 10 μL of the lipoxygenase solution.
Whole-genome re-sequencing
Sequencing libraries for Lox1 deficiency (IT160160 and IT276392) were constructed using a DNA sample preparation kit (Insilicogen Inc., Youngin, Republic of Korea). The DNA samples were subsequently sequenced on an Illumina NovaSeq 6000 platform, which generated paired-end reads of 151 base pairs. In the post-sequencing data processing pipeline, duplicate PCR reads were eliminated, followed by the removal of adapter sequences and low-quality bases using Trimmomatix-0.39 program (Bolger et al., 2014). The resulting clean reads were aligned to the soybean reference genome (Glyma.Wm82.a2.v1) (Schmutz et al., 2010) using BWA-MEM (Li, 2013), and subsequent SNP annotation was conducted based on gene location information provided by the soybean reference genome version 2 (SEEDERS Inc., Daejeon, Republic of Korea). Reads with more than 90% identical mapping were classified as ‘homozygous SNPs,’ while those with more than 40% but less than 60% variation in SNPs were classified as ‘heterozygous SNPs. ’ If read mapping was not feasible at a specific SNP locus, it was denoted with a lowercase ‘n and categorized as ‘N,’ indicating the presence of multiple mixed sequences.
DNA extraction and KASP assays
DNA was extracted from freshly harvested leaves of each F2:3 population plant using a DNeasy Plant Kit (Qiagen, Hilden, Germany). The DNA concentration and purity were measured using a Nanodrop ND 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Four KASP markers targeting the mutated region of the lipoxygenase gene were developed: two for Lox1 and one each for Lox2 and Lox3. The KASP genotyping was performed in a total volume of 10 μL, where 5 μL of DNA (at a concentration of 50 ng/5 μL) was combined with 0.14 μL of the KASP assay mix, and further supplemented with 5 μL of master mix. Genotyping was performed using the ABI StepOnePlus software (Applied Biosystems, Foster City, CA, USA).
The KASP PCR cycling conditions included a prereading stage at 30°C for 1 min, a holding stage at 94°C for 15 min, 10 touchdown cycles at 94°C for 20 s, followed by 35 cycles at 94°C for 20 s and 55°C for 1 min, with a final post-read stage at 30°C for 30 s. Endpoint fluorescence data analysis was performed using StepOne software (Applied Biosystems).
Statistical analysis
IBM SPSS Statistics 25 (IBM, Armonk, NY, USA) was used to perform a chi-square test (using α = 0.05 as the significance cutoff) to determine whether Mendel’s laws were followed.
RESULTS
Genetic locations of lipoxygenase genes
The genomic locations of soybean seed lipoxygenase genes were identified using Phytozome (https://www.phytozome.net/) (Fig. 1). Lox1 and Lox2 were found within the lipoxygenase domain of the NCBI soybean browser, specifically Glyma 13g347600 and Glyma13g347500, respectively, in a cluster spanning 41.50 kb. The distance between Lox1 and Lox2 was found to be only 3,000 bp (Fig. 1A). Lox3 (Glyma15g026300) is located on chromosome 15 within another lipoxygenase-rich gene cluster covering 23.54 kb and contains two distinct genes with lipoxygenase domains (Glyma15g026400 and Glyma 15g026500) (Fig. 1B). The Lox1, Lox2, and Lox3 gene models were determined to have sequences of 4,270, 4,297, and 4,351 bp from the start codon to the end codon, respectively. Lox1 consists of ten exons and 11 introns, whereas Lox2 and Lox3 have nine exons and eight introns, respectively.
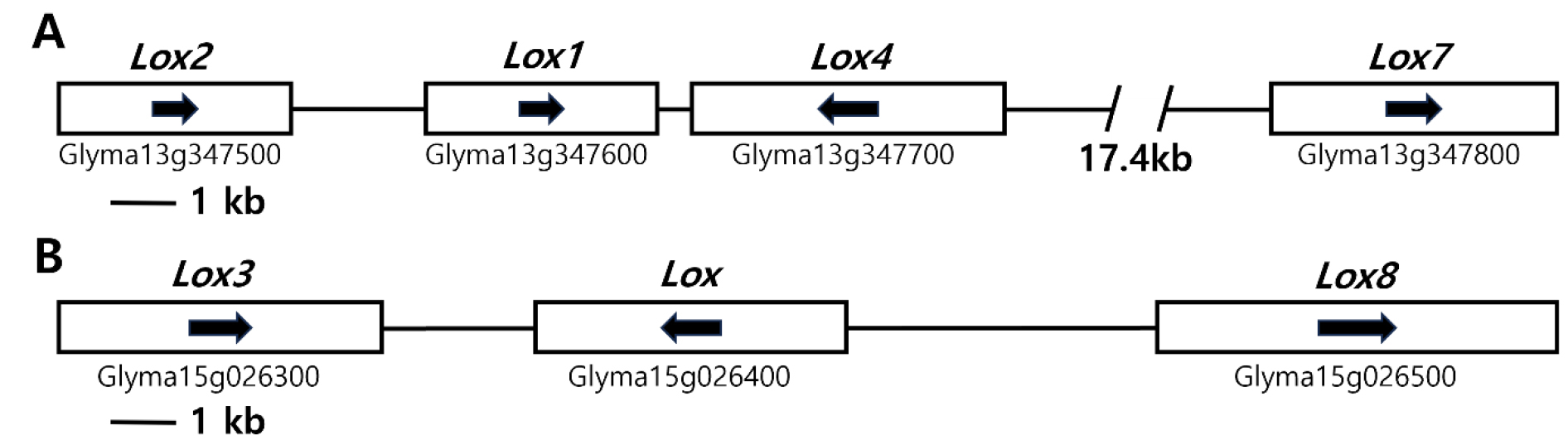
Fig. 1.
Lox gene sequences associated with the lipoxygenase of the seeds analyzed in this study in soybean genomes located on chromosomes 13 (A) and 15 (B). The gene clusters are depicted as white boxes, and the directionality of the genes is indicated by arrows within these boxes. The genetic nomenclature below the white box corresponds to the designations specified in the comprehensive soy genome assembly, which can be accessed at https://phytozome-next.jgi.doe.gov/info/Gmax_Wm82_a2_v1.
Analysis of lipoxygenase in 1,001 soybean accessions
To assess lipoxygenase levels, we conducted colorimetric analysis on a collection of 1,001 soybean accessions. Within this group, we identified two accessions (IT160160 and IT276392) that exhibited a deficiency solely in Lox1, and one (IT269984) that displayed a deficiency in both Lox1 and Lox2 (Table 1). The colorimetric method revealed that solutions lacking Lox1 and Lox2, which tested negative for lipoxygenase, retained a distinct blue color. In contrast, no cultivar lacking lipoxygenase was found for Lox3, as all test solutions became transparent, confirming the presence of Lox3 in all analyzed soybean accessions.
Table 1.
Results of lipoxygenase assay by colorimetric analysis in 1,001 soybean accessions.
| Accessions | Name | Origin | Lipoxygenase |
| IT160160 | L-1-5 | USA | Null lox1 |
| IT276392 | 1306 Black Manchurian | IDN | Null lox1 |
| IT269984 | Milyang 249 | KOR | Null lox1/lox2 |
Whole-genome re-sequencing
Re-sequencing analysis was conducted on two soybean accessions, IT160160 and IT276392, both of which exhibited Lox1 deficiency. The genome sequencing of these two accessions generated a combined total of over 428.45 million reads, equivalent to 64.70 Gb, with an average of approximately 214.23 million reads (32.35 Gb) per accession (Table 2). Following the cleaning process, 95.54% and 95.85% of the reads were successfully mapped to the reference soybean genome. The average sequencing depth ranged from 31.73x for IT160160 to 27.08x for IT276392 with an overall average depth of 29.41x.
A comparison of the two soybean accessions with the reference sequence revealed 3,720,664 variants, encompassing 3,096,358 SNPs and 624,306 indels. Specifically, IT160160 contained 1,368,642 SNPs, 264,096 indels, and 1,632,738 variants. In contrast, IT276392 contained 1,727,716 SNPs; 360,210 indels; and 2,087,926 variants.
Table 2.
Summary of genome re-sequencing and DNA sequence variants identified in IT160160 and IT276392.
Lox1 gene mutation
In IT160160, we identified a 74-bp deletion starting at position 2,752 within Lox1-a, which is consistent with the findings of a previous study (Fig. 2). This deletion triggered a premature stop codon, resulting in the formation of a truncated protein comprising 524 residues as opposed to the reference Lox1 protein, which contained 827 amino acid residues.
In IT276392, a mutation involving a change from C to A was detected at position 2,880 on chromosome 8. Interestingly, this mutation corresponds to the same SNP as Lox1-b, which was previously identified in another study. The C2880A mutation leads to a S568STOP alteration owing to a nonsense mutation in the start codon. Whole-genome sequencing revealed a C insertion at position 3,299 on chromosome 9 in both IT160160 and IT276392.
Detection of mutations in lipoxygenase genes using KASP markers
The KASP marker for Lox1-a (IT160160) was designed based on a 74-bp deletion spanning positions 2,752–2,825 of the related candidate gene, Glyma13g347600. Additionally, a KASP marker for Lox1-b (IT276392) was developed using a missense mutation at position 2,880 of Glyma13g347600. For Lox2, the KASP marker was designed around a missense mutation located in exon 8 of the related candidate gene, Glyma13g347500. For Lox3, a KASP marker was developed using the 101G indel located within exon 1 of Glyma15g026300 (Table 3).
Table 3.
List of four SNP markers for KASP analysis.
The Lox1-a KASP marker was successfully used to examine the genotypes of 169 F2 populations from a cross between Daepung 2 and IT160160. The results showed a clear separation of the three alleles: 35 wild types, 89 heterozygotes, and 45 mutants (Fig. 3A and Table 4). We found that the KASP analysis of Lox1-a showed 96% concordance with the colorimetric assay (Table 4 and Suppl. Table 1). The Lox1-b KASP marker was used to successfully genotype 147 F2 populations from Daepung 2 × IT276392. Of these, 48 were wild-type, 76 were heterozygous, and 23 were mutant (Fig. 3B and Table 4). Although the 3:1 ratio was not strictly maintained, the concordance rate between KASP and colorimetric assays was 96% in the Daepung 2 × IT160160 population. Consequently, although the separation rate results of the KASP marker and colorimetric analysis were consistent, only the KASP marker could differentiate between heterozygous enzymes, resulting in significantly more accurate outcomes (Table 4).
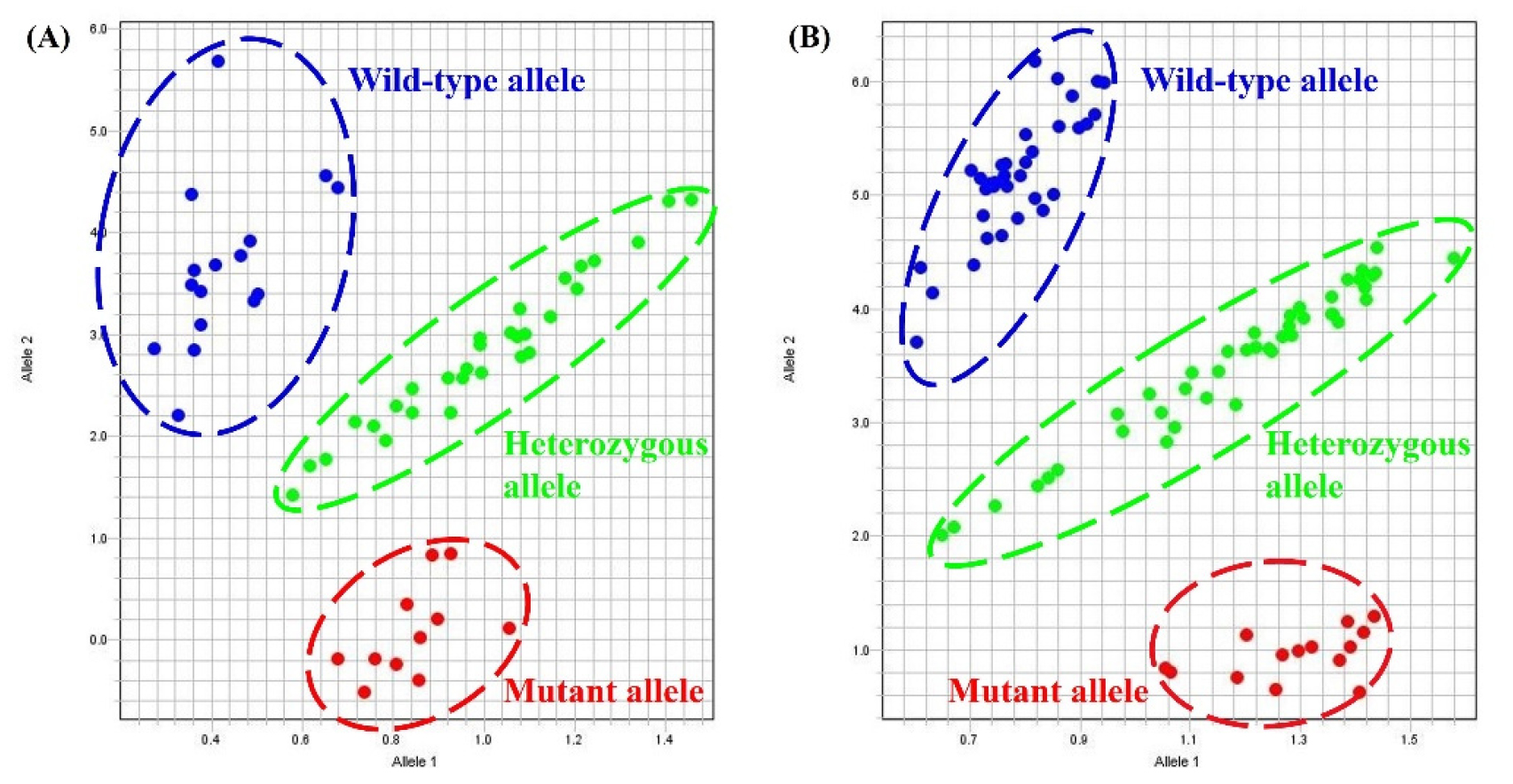
Fig. 3.
Genotyping results of the Lox1 KASP assays. (A) Three distinct clusters were revealed using the Lox1-a KASP marker in the 169 F2 plants derived from Daepung 2 × IT160160. (B) Three distinct clusters were revealed using the Lox1-b KASP marker in the 147 F2 plants derived from Daepung 2 × IT276392.
Table 4.
KASP results from two different F2:3 populations.
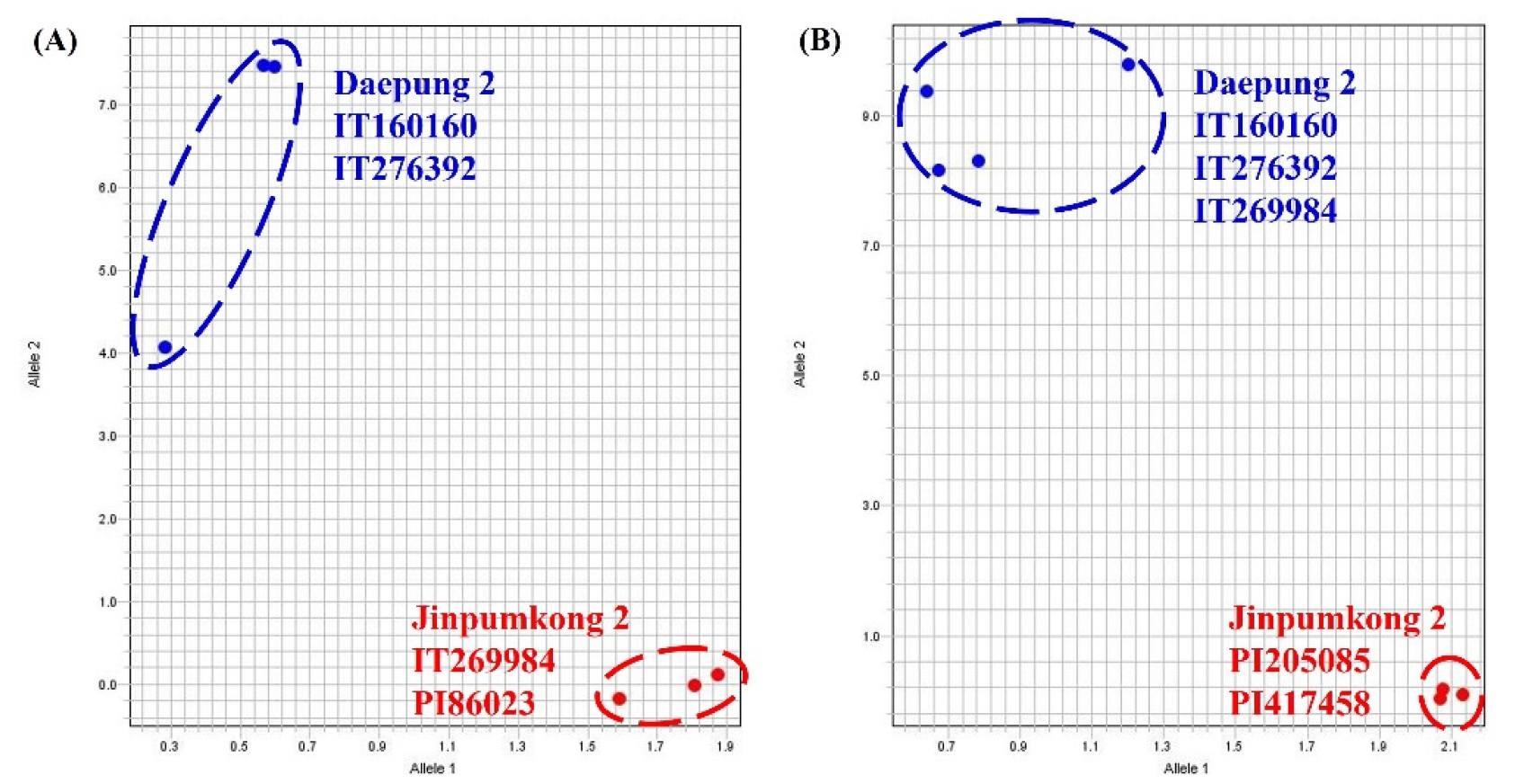
To validate the KASP marker for null-Lox2, we tested Daepung 2, IT160160, IT276392, and IT269984 using two cultivars previously identified as Lox2-deficient in other studies (Jinpumkong 2 and PI86023). The results confirmed that Daepung 2, IT160160, and IT276392 were wild type, whereas IT269984, Jinpumkong 2, and PI86023 exhibited the same null Lox2 mutant type (Fig. 4A). To validate the KASP marker for null-lox3, we compared Daepung 2, IT160160, IT276392, and IT269984 with Jinpumkong 2, PI205085, and PI417458, which have been shown to exhibit LOX3 deficiency in previous studies. Consequently, it was confirmed that Daepung 2, IT160160, IT276392, and IT269984 were wild-type, whereas Jinpumkong 2, PI205085, and PI417458 were mutant types (Fig. 4B). These findings suggest that the KASP markers developed in this study can rapidly and accurately determine the presence or absence of Lox1, Lox2, and Lox3 deficiencies.
DISCUSSION
Soybean lipoxygenases have received significant attention because they produce oxidized compounds that contribute to unpleasant flavors in soy products (Kitamura, 1984). Development of Lox-deficient soybean varieties requires precise evaluation assays to select suitable genotypes. Currently, colorimetric analysis is the most widely used technique for this purpose, because it provides a precise and reliable assessment of the absence of Lox. This method is based on enzyme-substrate reactions and detects the oxidation of a reagent by a specific lipoxygenase isoenzyme, depending on the pH of the test solution in which the isozyme exhibits optimal activity (Mack et al., 1987). In the presence of the isozyme, linoleic acid undergoes oxidation, resulting in the reduction of Lox1 and Lox2, whereas Lox3 undergoes oxidation and becomes transparent, facilitating the confirmation of lipoxygenase deficiency. Our preliminary experiments indicated that the most distinct differences in the presence of Lox could be observed within a shorter period, which led us to adopt this method in our study. However, colorimetric analysis has three limitations. First, the duration for which seeds are allowed to stand after grinding significantly affects the accuracy of colorimetric analysis. Testing ground and aged seeds often yields inconsistent results due to decreased enzyme activity. Second, beans with black seed coats posed challenges in visual distinction after being mixed with the solution, necessitating the removal of the seed coats before conducting the experiment. Finally, colorimetric analysis could not differentiate between the heterozygotes. However, the use of molecular markers has enabled clear differentiation between wild types, heterozygotes, and mutants. This approach not only overcomes the limitations of colorimetric analysis, but also underscores the utility of KASP markers in efficiently identifying and selecting soybean cultivars lacking lipoxygenases, thereby enhancing soybean breeding programs aimed at improving food quality.
Through colorimetric assay using 1,001 soybean accessions, we identified two soybean accessions lacking Lox1 (IT160160 and IT276392), and one soybean accession lacking both Lox1 and Lox2 (IT269984). IT160160, which was collected in the USA and formerly known as L-1-5, has been reported to exhibit a null Lox1 phenotype (Davies & Nielsen, 1986). In this study, we confirmed that IT160160 exhibited a 74-bp deletion starting at position 2,752 within Lox1 (Glyma13g347600), which is the same as PI408251 (Kitamura, 1984). Furthermore, our investigation revealed similarities between the mutations identified in IT276392 and PI133226, which were introduced in Indonesia in 1939. Both IT276392 and PI133226 were originally sourced from Indonesia, indicating a potential genetic kinship likely attributable to their shared geographical origins. IT269984 was developed by crossbreeding Daemang and Jinpumkong 2. Consequently, the use of Jinpumkong 2 as a parent variety resulted in the discovery of a unique double deficiency characterized by the absence of both Lox1 and Lox2 traits within IT269984. Additionally, no resources with natural triple lox deficiency could be identified. Previous studies have shown that varieties such as Jinpumkong 2, OX948, and H70 exhibit null Lox3 (Kim et al., 1997; Lee et al., 2014; Reinprecht et al., 2011). OX948 and Jinpumkong 2 share a common feature: they possess a Lox3-a allele characterized by a frameshift mutation in exon 1, which introduces a stop codon at position 101 and leads to premature termination of the protein (Lenis et al., 2010). However, in the case of H70, a triple-null Lox variety created through gamma irradiation, a distinct single point mutation (A-G) was identified in exon 6 of Lox3 (Lee et al., 2014). This mutation changes amino acid 405 from histidine to arginine, rendering Lox3 inactive (Lee et al., 2014). Consequently, unlike Lox1 and Lox2, Lox3 mutations exhibit a greater diversity in their genetic alterations. Therefore, triple-deficient resources must be artificially created using methods such as gamma irradiation.
In the Korean soybean accessions, two mutations were identified in different Lox genes: a 74-bp deletion in exon 8 of Lox1-a and a C-A missense mutation in exon 8 of Lox1-b. Jinpumkong 2 harbors a T-A mutation in exon 8 of Lox2 and a 101G indel in exon 1 of Lox3. These genetic variations led to the development of a targeted KASP marker that utilizes the identified genetic variations for detection. Previous studies have devised simple probes and KASP markers targeting specific genetic variations within various accessions, such as Lox1-a in PI408251 (74-bp deletion), Lox2 in PI86023 and Jinpumkong 2, and Lox3 mutations in Jinpumkong 2, PI205085, and PI417458 (Lenis et al., 2010; Patil et al., 2017). These markers enabled discrimination between wild-type and mutant alleles by analyzing distinct peak profiles. In the present study, we developed a novel KASP marker, marking the first instance of its use by focusing on the mutant SNP present in PI133226 (Kitamura, 1984). In the case of Lox1, we compared the colorimetric analysis with the KASP analysis using the F2:3 population created by crossing Daepung 2 (wild-type lipoxygenase) with IT160160 and IT276392 (null Lox1). The results showed a substantial agreement of 96% between colorimetric analysis and the KASP marker, underscoring the effectiveness of the KASP marker in identifying Lox gene mutations in our study.
Therefore, we developed comprehensive KASP markers for each of the four soybean lox genes. KASP analysis exhibited high concordance between genotypes and phenotypes, validating the robustness of our analysis. This approach not only overcomes the limitations of colorimetric analysis but also highlights the effectiveness of KASP markers in the efficient identification and selection of soybean varieties lacking lipoxygenase. This will contribute to the enhancement of soybean breeding programs aimed at improving food quality.